Superfluid breaks the STAR bottleneck with Tracer and NVIDIA Parabricks
Superfluid Dx partnered with Tracer to benchmark CPU vs GPU-accelerated STAR alignment for Alzheimer's research. Results show 67% faster runtime at near cost parity with NVIDIA L40S GPUs.
const metadata = ;
Superfluid Dx develops cf-mRNA-based diagnostic models for Alzheimer's disease. Their workflows rely on accurate splice-junction detection, which makes the STAR alignment tool a critical stage in every pipeline. Using Tracer, who provides kernel-level observability on cloud instances, they found that the STAR tool was the main bottleneck, both in runtime duration and cost and wanted to accelerate this pipeline step.
To understand whether GPUs could meaningfully accelerate alignment, Superfluid Dx partnered with Tracer and leveraged NVIDIA Parabricks to benchmark CPU-optimized runs against GPU-accelerated STAR across multiple dataset sizes. The results outline a predictable scaling pattern and a clear direction for future optimization.
Executive summary
Using Tracer's kernel-level telemetry, Superfluid Dx evaluated a Parabricks-powered GPU approach to understand potential gains in speed, cost efficiency, and scalability.
Key GPU Parabricks findings:
- NVIDIA L40S delivered ~67% faster runtime at near cost parity with CPU for datasets with ~110M reads
- GPU-accelerated STAR alignment reduced runtime by approximately 50% on test datasets compared to the CPU baseline
- GPU speedups increased more as dataset size increases, pointing toward a future crossover point where GPUs become the default
How STAR alignment is used in Alzheimer's research
Superfluid Dx uses two-pass STAR alignment to maximize splice-junction accuracy, an essential requirement when studying RNA isoforms and splicing changes implicated in early neurodegeneration.
Two-pass alignment improves interpretability but adds significant compute load, pushing memory, I/O, and parallelization limits. For workflows that run hundreds of millions of reads per sample, even small inefficiencies compound into hours of lost compute. Any meaningful acceleration of the STAR step accelerates downstream scientific discovery.
CPU Baseline: What Superfluid Dx uncovered with Tracer
Tracer's kernel-level signals revealed that:
- STAR used only ~24% of CPU due to network and disk I/O stalls
- Reads and writes were the limiting factor, not compute
- Moving to modern instances with faster storage and networking unlocked a 7x speedup
This CPU-optimized baseline served as the reference point for GPU benchmarking.
Benchmarking GPU acceleration with Parabricks
Superfluid Dx evaluated CPU vs. GPU performance across:
- Test dataset (~20M reads)
- Production sample (~110M reads)
Hardware included NVIDIA A10 Tensor Core GPU, NVIDIA L40S, and NVIDIA A100 Tensor Core GPU-based AWS instances.
Phase 1: Test Dataset (~20M reads)
Goal:
Validate GPU alignment and understand initialization overhead.
Findings:
- Parabricks alignment reduced STAR runtime duration by ~50% and reduced costs by 44% for the best performing setup
- Best performance was achieved on a single GPU, as multi-GPU splitting introduced overhead and slowed small workloads
- The --low-memory parameter needs to be included to manage smaller memory. By default, Parabricks only supports GPUs above 16 GB. To support smaller GPUs (T4 only has 16 GB available), the parameter needs to be included
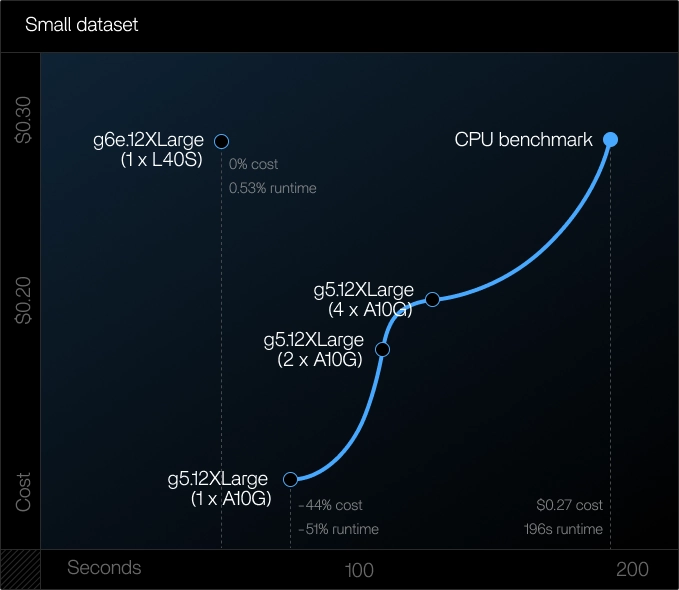
Learnings in phase 1:
For small datasets, GPUs deliver both faster and cheaper alignment, especially when a single GPU has enough memory to avoid file splitting.
Phase 2: Production sample size (~110M reads)
Goal:
Understand scaling behavior, GPU behaviour under production workloads and cost efficiency.
Findings:
- Runtime duration improved by ~67% vs CPU. GPU speedups as a % vs CPU set increased with dataset size, which indicates better scale benefits at higher read counts
- Cost approached parity. NVIDIA L40S instances runs landed within ~3% of the CPU cost, while delivering significantly faster throughput
- Larger instances did not provide sufficient speed improvements to offset the GPU cost. NVIDIA A100 showed a ~5% cost increase vs CPU baseline on the medium dataset
- Parameter tuning made a measurable difference. Enabling --num-threads 128, --gpu-write, --gpu-sort, and --memory-limit=1000 unlocked additional performance on A100 and L40S by fully utilizing 128 CPU threads, offloading sort and write paths to the GPU, and lifting the RAM ceiling to 1 TB to avoid memory-bound execution
- Multi-GPUs did not improve alignment speed. Splitting medium datasets across multiple GPUs resulted in longer runtime due to merging overhead
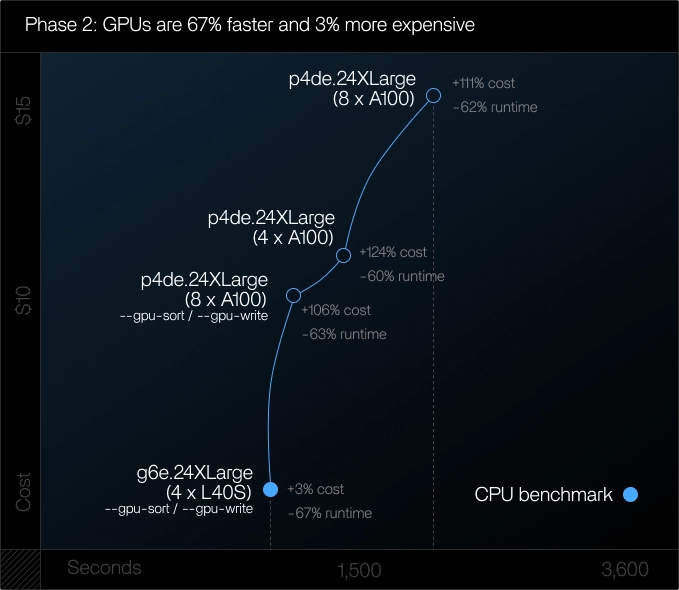
Learnings in phase 2:
For datasets at c.110M reads, L40S GPUs emerged as the "almost-crossover" option, nearing CPU cost with dramatically faster execution.
The potential for larger datasets
Overall, the bigger the dataset, the larger the relative speed improvement (in %), compared to the CPU benchmark. For datasets at +150M reads, it requires further conversations with sales teams at cloud providers to use GPUs with the extreme memory necessary. This should however not be an impossible problem to have.
The ROI: When acceleration pays for itself
Across all experiments, one pattern held: The larger the dataset, the more NVIDIA Parabricks outperform CPU like-for-like comparison. Parabricks speedups compress time-to-science, reduce hypothesis testing backlog, and enable faster iteration cycles, which is critical in research settings where teams are forced to analyse hypotheses daily, not weekly. Finally, more modern, exclusive GPUs are expected to generate even better results due to faster I/O and memory technology.
Tracer evaluated Parabricks by exposing I/O stalls, memory ceilings, and underutilization before they caused wasted runs or misleading results. The evaluation also weighed different options in terms of costs and speed, depending on the need and lifecycle of the company.
Benchmark your alignment step
Many pipelines still assume CPUs are the default, this work shows that for STAR-heavy RNA-seq workloads, NVIDIA Parabricks tools are quickly becoming the more compelling choice.
- Small datasets: GPUs are faster and often cheaper
- Medium datasets: L40S delivers near cost parity with a 67% speed improvement
For Alzheimer's research, where alignment accuracy directly influences downstream model quality, every speed improvement accelerates discovery.
Get started with Tracer
Using Tracer's kernel-level observability platform, this study found that STAR was disk I/O-bound, not compute-bound, and CPU utilization was limited to ~24%, providing an opportunity to switch to lower cost, modern instances with significantly higher data sample throughput.
Tracer's kernel-level observability solution gives full pipeline and infrastructure visibility so that you can (1) identify better instances for your workloads, (2) reduce cloud costs with better provisioning, (3) speed up pipelines without crashes, and (4) debug faster with all cloud and pipeline logs in one place. Tracer requires no code changes, a single command-line to implement, and works with any bioinformatics or data engineering framework.
About Superfluid Dx
Superfluid is developing the first high-performance, predictive blood-based test for Alzheimer's Disease (AD) and related dementias that directly assays mRNA transcripts from the brain via its platform technology of cell-free messenger RNA (cf-mRNA) analysis and machine learning. This next-generation novel liquid biopsy technology enables non-invasive measurement of the dynamic biology of organs throughout the body, including the brain. Our precise understanding of the underlying pathways of disease will transform AD disease care and treatment.
Superfluid is led by a small, highly accomplished team, including founder Steve Quake (Stanford Professor and Head of Science at CZI) and CEO Gajus Worthington (former founder and CEO of Fluidigm). The company has published extensively in peer-reviewed journals and is well funded by notable investors, including Brook Byers and Reid Hoffman. Superfluid is also supported by the National Institutes of Health and the Alzheimer's Drug Discovery Foundation.
Download Parabricks today
Download [NVIDIA Parabricks](https://catalog.ngc.nvidia.com/orgs/nvidia/teams/clara/containers/clara-parabricks?nvid=nv-int-tblg-737311-vt12) to get started with GPU-accelerated genomic analysis and join the conversation on the [NVIDIA Parabricks Developer Forum](https://forums.developer.nvidia.com/c/healthcare/parabricks/290).

