The Silent Gene Name Corruption Everyone Misses
A Simple and Practical Guide to Stop Excel From Breaking Your Biology
const metadata = ;
Summary
Gene name corruption happens when Microsoft Excel automatically converts gene symbols into dates or numbers. This silent conversion damages datasets, breaks pipelines, and lowers scientific reproducibility. Even today, more than 30% of publications that include gene lists show errors caused by Excel. This article explains why this happens, how to avoid it, how to detect errors early, and how to implement safe workflows. Real scientific figures and tables show how widespread this problem is across journals, species, and data types. You will also learn about masked genomic regions, random genomic sites, QC checks, best practices, and code snippets for detecting corruption. This is a complete, practical guide to protecting your data from Excel.
Introduction
If you work in bioinformatics, you probably know the feeling: someone sends you an Excel file with gene names, you open it, and suddenly SEPT9 becomes 9-Sep, MARCH1 becomes 1-Mar, and DEC1 becomes 1-Dec. Excel does this automatically, without asking or warning.
This problem is not new. It is silent data corruption, and it has affected genomics research for more than a decade. Once Excel converts a gene name and the file is saved, the original gene symbol is permanently lost.
This article explains why this happens, what it breaks, and how to stop it. It includes scientific evidence, code examples, and practical steps you can use immediately. You will also learn why masked genome regions and random genomic sites become even more dangerous when corrupted data reaches downstream pipelines.
Key Takeaways
- Excel automatically converts some gene names into dates.
- Once saved, the corruption is permanent.
- Prefer .csv, .tsv, or .txt instead of Excel.
- If Excel must be used, import gene columns as text.
- Ensembl and RefSeq IDs never convert into dates.
- QC scripts can detect corrupted names before analysis.
- More than 30% of published gene lists contain Excel errors.
- These errors harm reproducibility and biological interpretation.
- This article includes figures, tables, and code to help prevent the issue.
- Educating collaborators is necessary to avoid future mistakes.
Glossary
- HUGO Gene Symbols: Short gene names like SEPT9 and MARCH1.
- Masked Regions (NNNs): Genome areas with unknown bases represented as “N.”
- Random Genomic Sites: Random genomic positions used for controls or background distributions.
- Silent Data Corruption: A change that happens without warning.
- Stable Identifiers: IDs such as Ensembl or RefSeq that never resemble dates.
- QC Checks: Steps to ensure accuracy before running pipelines.
- Auto-formatting: Excel’s automatic attempt to interpret text as dates or numbers.
Random Genomic Sites Explained
Random genomic sites are used in enrichment tests, motif analysis, and background modeling. If metadata or gene names associated with these positions become corrupted, entire statistical comparisons become unreliable. This amplifies Excel-based mistakes and leads to false biological conclusions.
Masked (NNN) Regions Explained
Masked regions contain uncertain bases. These appear as “N” in FASTA files. Misalignment, coordinate corruption, or altered identifiers caused by Excel can produce larger downstream errors when such regions are involved. This is another reason why upstream data integrity must be maintained.
The Problem: Excel Silently Changes Gene Names
Excel automatically formats anything that resembles a date. Here are common examples:
Original Gene
Excel Output
SEPT9
9-Sep
MARCH1
1-Mar
DEC1
1-Dec
NOV1
1-Nov
FER
12-Feb
Excel does not notify you. It simply converts the value.
Why this happens:
- Excel tries to “help” users by interpreting patterns.
- HUGO symbols overlap with month abbreviations.
- Many biologists still use Excel for convenience.
- Saving the file makes the corruption irreversible.
A real Biostars post reads, “SEPT9 became sept-9 after storing gene annotation in Excel.”
How Common Is This Problem? Scientific Evidence
Below are real figures from open-access scientific studies that show how widespread this problem is.
Figure 1: Publications Affected by Excel Gene Name Errors (2014–2020)
This figure shows how many scientific publications contained Excel-induced gene name errors between 2014 and 2020. It highlights that the problem is widespread and persistent, with more than 30% of papers showing corrupted gene lists every year.
.png)
Source: PLOS Computational Biology (CC-BY 4.0)
Figure 2: Error Rates Across Journals and Years
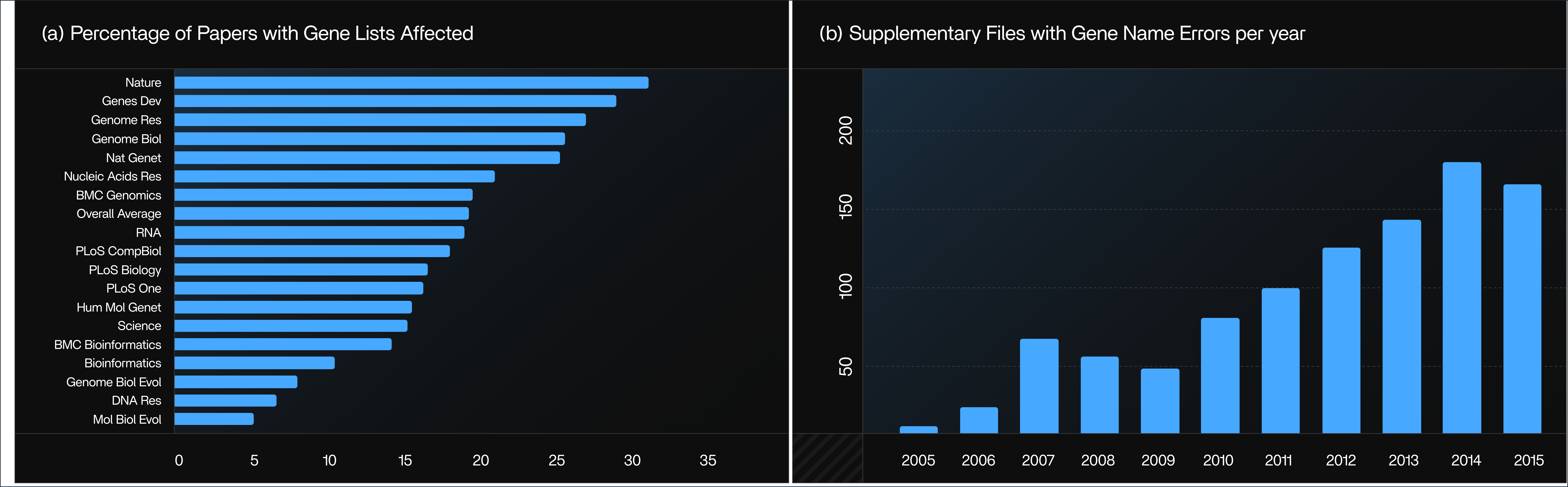
Source: Genome Biology (CC-BY 4.0)
High-impact journals such as Nature, Genes & Development, and Genome Research have some of the highest error rates. This figure compares error rates across different journals and publication years. It shows that even high-impact journals consistently publish gene lists affected by Excel’s automatic date conversion.
Figure 3: Journal-Specific Growth of Excel errors
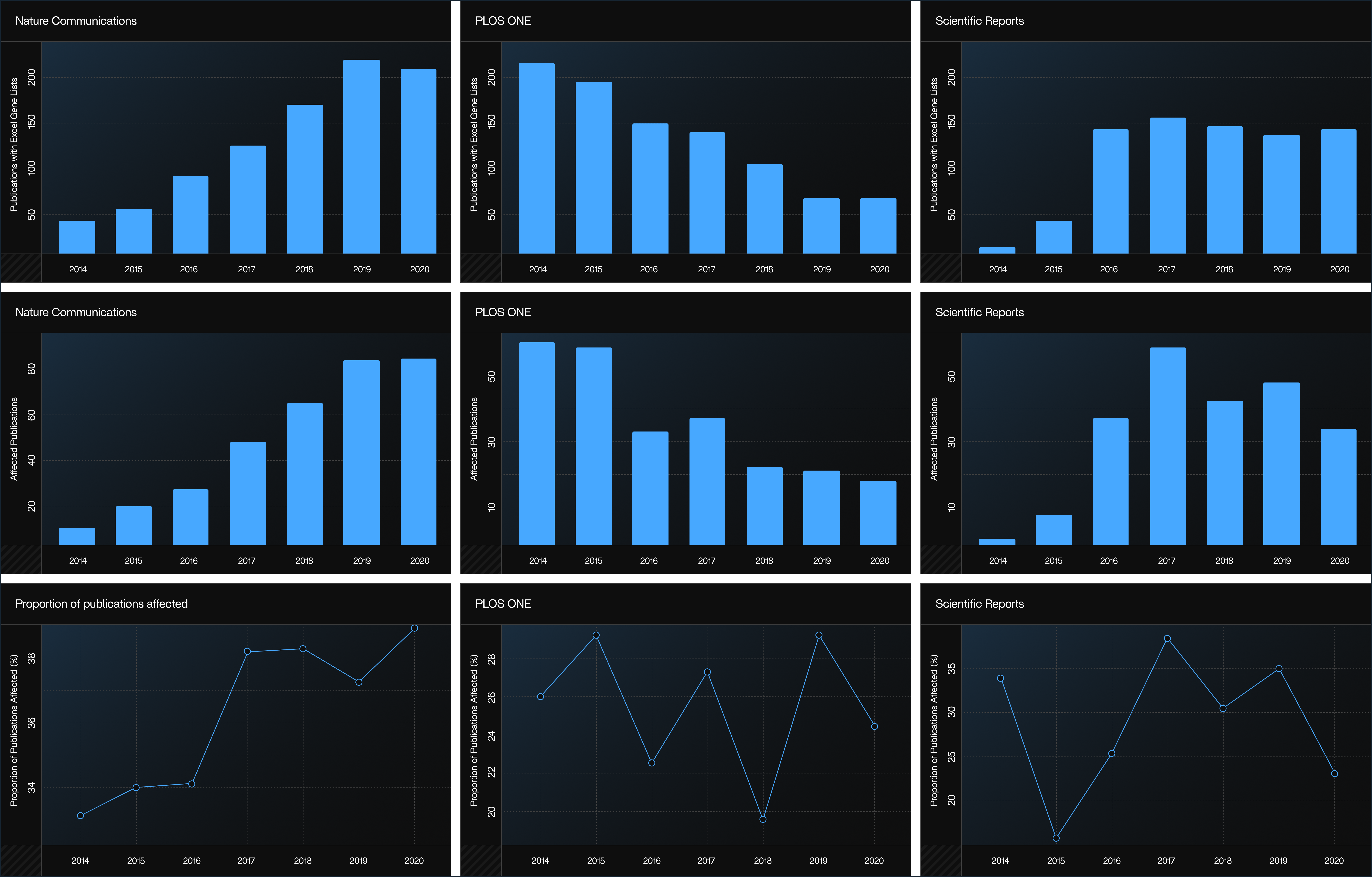
Source: PLOS Computational Biology (CC-BY 4.0)
Even top journals like Nature Communications, Scientific Reports, and PLOS ONE show frequent corruption. This figure illustrates how specific journals such as Nature Communications, PLOS ONE, and Scientific Reports experience yearly increases in Excel-related gene name errors. It reveals that the issue is not limited to low-quality journals but affects top-tier publications as well.
Real-World Evidence of Excel Gene Name Corruption in Databases
[PLACEHOLDER FIGURE 4]
Figure 4. Excel-Corrupted Gene Name Appearing in an NCBI LocusLink Record
This figure shows how a gene symbol such as SEPT2 is incorrectly displayed as “2-Sep” in the NCBI LocusLink database, demonstrating how Excel-induced date conversion can propagate into public biological databases. Source: BMC Bioinformatics (CC-BY).
[PLACEHOLDER FIGURE 5]
Figure 5. Corrupted Gene Symbol in the Human–Mouse Homology Map
This figure illustrates how the same Excel-converted gene name (“2-Sep”) appears in cross-species homology maps, showing how silent corruption affects comparative genomics and downstream biological interpretation. Source: BMC Bioinformatics (CC-BY).
This figure illustrates how the same Excel-converted gene name (“2-Sep”) appears in cross-species homology maps, showing how silent corruption affects comparative genomics and downstream biological interpretation. Source: BMC Bioinformatics (CC-BY).
Additional Scientific Tables Supporting the Problem
Table 1: Software That Converts Gene Names Automatically
Software
Microsoft Excel
Google Sheets
LibreOffice
Gnumeric
Text file open
Yes
Yes
No
No
Pasting data
Yes
Yes
No
No
Typing
Yes
Yes
No
No
Link: https://doi.org/10.1371/journal.pcompbiol.1008984.t001
Microsoft Excel and Google Sheets convert gene names on opening, pasting, and typing. LibreOffice and Gnumeric do not.
Table 2: Taxa and Genes Affected by Excel Conversion
Taxa
Genes
Genes affected
Taxa affected
Vertebrates
310
5,263,175
1,325
76
Metazoa
59
525,867
17
3
Plants
60
244,101
35
4
Fungi
59
788,221
140
12
Protists
39
163,026
27
9
Link: https://doi.org/10.1371/journal.pcompbiol.1008984.t002
Species from fungi to vertebrates experience gene name corruption.
Table 3: Publications Screened, Excel Files Screened, and Affected Files
2014
2015
2016
2017
2018
2019
2020
Total
Publications screened
19976
21204
22261
23976
24986
26046
27690
166139
Excel files screened
2948
4318
4472
4355
4824
5481
6443
32841
Excel files with gene lists
2286
3037
3331
3021
3566
3342
4496
23670
Publications with Excel gene lists
936
1491
1579
1412
1653
1823
2223
11117
Publications with suspected gene name errors
284
490
477
443
475
594
707
3470
False positive Excel files
8
0
7
5
15
4
11
50
False positive publications
2
0
6
3
11
3
9
34
Affected Excel files
429
701
653
648
703
914
1038
5086
Affected publications
282
490
471
440
464
591
698
3436
Proportion of publications affected (%)
30.1%
32.9%
29.8%
31.2%
28.1%
32.4%
31.4%
30.9%
Link: https://doi.org/10.1371/journal.pcompbiol.1008984.t003
More than 166,000 publications and 32,000 Excel files were examined.
Table 4: Species-Wise Impact on Publications
Species
Publications with Excel gene lists
Affected publications
Proportion of publications affected
M. musculus
1577
609
38.6%
H. sapiens
7936
2419
30.5%
C. elegans
124
31
25.0%
D. melanogaster
607
142
23.4%
S. cerevisiae
443
93
21.0%
R. norvegicus
327
68
20.8%
D. rerio
251
48
19.1%
A. thaliana
511
76
14.9%
G. gallus
1827
172
9.4%
O. sativa
10
0
0.0%
Link: https://doi.org/10.1371/journal.pcompbiol.1008984.t004
Human and mouse datasets have the highest rate of corruption.
Table 5: Journal-Level Error Statistics
Journal name as it appears in PMC
Number of articles with Excel gene lists
Number of affected articles
Proportion of articles affected (%)
Nat Commun92034537.5%
PLoS One94624425.8%
Sci Rep76722729.6%
BMC Genomics66016625.2%
PLoS Genet44813429.9%
Oncotarget32610732.8%
Front Genet3139430.0%
eLife2438936.6%
Proc Natl Acad Sci USA1557347.1%
Cell Rep1587144.9%
Genome Biol1936634.2%
Nature1185244.1%
Nat Genet1404834.3%
Genome Med1374432.1%
PeerJ1373928.5%
Cell743952.7%
Clin Epigenetics1093834.9%
Nucleic Acids Res1203630.0%
BMC Med Genomics1173126.5%
Front Oncol853136.5%
Transl Psychiatry732939.7%
BMC Cancer1052826.7%
PLoS Pathog802733.8%
Commun Biol742736.5%
PLoS Biol662639.4%
Aging562646.4%
EBioMedicine512651.0%
Epigenetics Chromatin642539.1%
PLoS Comput Biol972424.7%
Oncogene532241.5%
iScience582034.5%
Sci Adv562035.7%
BMC Bioinformatics771924.7%
G3741520.3%
Hum Mol Genet531528.3%
BMC Plant Biol52611.5%
Front Plant Sci7556.7%
Link: https://doi.org/10.1371/journal.pcompbiol.1008984.t005
Some journals show error rates above 50%, meaning more than half their gene lists are corrupted.
Why This Matters: The Damage It Causes
1. Corrupted gene lists
When Excel converts gene names into dates, downstream tools can no longer recognize them. This means those genes are treated as missing, leading to incomplete annotations and datasets that silently lose important biological information.
2. Broken workflows
Excel-induced errors disrupt entire analysis pipelines. Tools like DESeq2, edgeR, and pathway enrichment packages may fail, drop corrupted entries, or produce incomplete results because key genes no longer match reference databases.
3. Reproducibility issues
Once gene names are altered, other researchers cannot reproduce your results, even if they use the same pipeline. Hidden differences in input files lead to diverging outputs, weakening scientific reliability and trust.
4. Incorrect biological conclusions
Missing or corrupted genes can shift pathway interpretations, hide regulatory relationships, or distort disease-related findings. In some cases, the biological story changes completely because essential genes vanish from the dataset.
Best Practices to Avoid Gene Name Corruption
1. Avoid Excel entirely
Using .txt, .csv, or .tsv files prevents automatic formatting and keeps your gene names exactly as they are. These formats are also easier to track, automate, and integrate into pipelines.
2. If Excel must be used
Always import gene columns as Text using “Data → From Text/CSV → Column Type: Text.” This stops Excel from converting gene symbols into dates or numbers.
3. Prefer stable identifiers
Ensembl or RefSeq IDs are safer alternatives because they never resemble dates, ensuring they remain unchanged across tools, pipelines, and file formats.
4. Run QC scripts before analysis
Simple Python, R, or Bash scripts can detect corrupted values early. Running these checks before every pipeline run helps catch silent errors before they break the analysis.
QC Code Examples
Python: Detect Excel Conversions
`bash
import pandas as pd
def detect_excel_corruption(file):
df = pd.read_csv(file)
corrupted = df[df.iloc[:,0].astype(str).str.contains(r'\d-[A-Za-z]', na=False)]
return corrupted
print(detect_excel_corruption("genes.csv"))
`
Bash: Quick Terminal Scan
`bash
grep -E "^[0-9]-[A-Za-z]$" genes.txt
`
R: Identify Corrupted Names
`bash
genes <- read.csv("genes.csv", stringsAsFactors = FALSE)
pattern <- "^[0-9]-[A-Za-z]$"
print(genes[grepl(pattern, genes$Gene), ])
`
Python: Restore Gene Names
`bash
mapping =
df['Gene'] = df['Gene'].replace(mapping)
`
Pipeline Safety Check
`python
def validate_before_pipeline(file):
df = pd.read_csv(file)
if df.iloc[:,0].str.contains(r'\d-[A-Za-z]').any():
raise ValueError("Pipeline blocked: Corrupted gene names detected.")
`
Before vs After Excel Conversion
`bash
Before:
SEPT9
MARCH1
DEC1
After:
9-Sep
1-Mar
1-Dec
`
Checklist Before Running Analysis
- Never store or share gene lists in Excel
- Always use .csv or .tsv
- Run corruption detection scripts
- Use stable identifiers
- Keep raw backups
- Confirm collaborators did not open your files in Excel
- Validate data formats before pipelines
Conclusion
Excel continues to corrupt gene names by automatically converting them into dates, even in 2025. This problem affects publications, species, pipelines, and entire workflows. However, with simple steps, using safe file formats, applying QC scripts, preferring stable identifiers, and teaching collaborators, you can eliminate this issue permanently. Clean data leads to clean science. Protecting your gene names is the first step.

